Genetic Algorithm for Tile Self Assembly
In the fall of 2011 and spring of 2012, four Computer Science students – Adam Smith, Jaris Van Maanen, Joel Gawarecki and Linsey Williams, studied tile self-assembly and the TAS software system developed at ISU. Tile self-assembly systems serve as models of DNA molecules designed to act as four-sided building units that can self-assemble to various shapes. Research on the design of such nanoscale constructs has shown their high potential usefulness in the area of nanotechnology. Given a target shape of tiles, the goal is to find the tile set that will self-assemble in the target shape.
The students designed and implemented a distributed genetic algorithm for evolving tile sets to form a pre-determined shape. The input to our genetic algorithm is a collection of randomly constructed tile sets. The output of the genetic algorithm is a set of tiles that most closely assembles to the target shape. The genetic algorithm was implemented in a distributed computational architecture used to speed up the process of obtaining subsequent generations of tile sets.
The chosen representation of the tile sets, the experiments with selection, cross-over, and mutation methods, and the distributed implementation of the genetic algorithm were described in a paper presented at the Midwest Instruction and Computing Symposium April 13 – 15, 2012, at UNI. They also presented at the Simpson College Undergraduate Symposium (April 19, 2012). The students were supervised by PI Lydia Sinapova.
Imaging and Characterization of DNA nanostructures
A 50 micro liter batch of the triangles was brought back to Simpson College where Jesse Smiles and Arlene Ford (in conjunction with Ron Warnet, Patricia Singer and Stephen Henrich) have been using Simpson College’s Atomic Force Microscope to image and characterize these 2D triangle shapes, so that the atomic force microscope could be optimized for the analysis of the DNA nano-strucutres. The solution of origami particles was diluted by 10x with distilled water and was then placed on a cleaned mica surface to dry under desiccated /dried air. This mica was then loaded into the atomic force microscope and imaged using its alternating contact mode. A common set point of 55% was used and images were obtained of the 300 nm sized triangles at a scale of 4 nm x 4 nm.
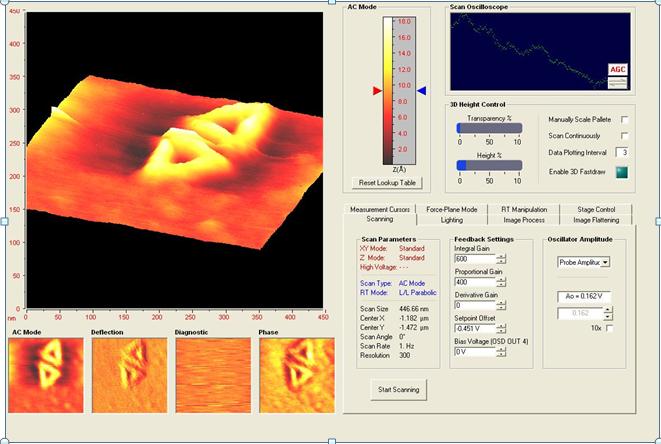
AFM Images of 2D nanoscale triangles captured at Simpson College
Exploring Drug delivery through DNA origami
Stephen Henrich and Jesse Smiles (both Chemistry students) examined a DNA origami vehicle with the capability of interfacing with cells in vitro, triggering programmed release of a molecular payload in response to cellular cues to stimulate intracellular signaling. Their research was focused on developing this model to yield a construct with high efficacy. They have used caDNAno to design origami structures for this purpose, and atomic force microscopy to conduct imaging for characterization of these nanostructures. They presented a poster “Drug delivery and DNA Origami: Rational design for targeted, intracellular delivery of molecular payloads” at the Simpson College Undergraduate Symposium (April 19, 2012). These students were supervised by Arlene Ford, Pat Singer and Ron Warnet.
DNA Origami Research
Students at Simpson College have been actively engaged in designing a novel DNA origami structure using CaDNAno software over the Fall of 2012. This structure will enable testing of a variety of binding strategies for linking DNA tiles together into larger self-assembled structures We obtained both the M13 scaffold DNA and 37 separate synthetic DNA staple strands to produce our DNA origami structure. All materials were taken to Iowa State University for the initial assembly of the DNA origami structure. The November 2012 visit was led by Arlene Ford (physicist) and Derek Lyons (biochemist), who were accompanied by six students: Jesse Smiles, Courntey Muhlbauer, Joe Grimely, Maia Kelly, Andrew Dexter, and Peter Reitgraf. Our group met with Divita Mather, a graduate student in Eric Henderson’s laboratory, to refine our techniques in all steps in DNA origami design, construction, and visualization. Under the guidance of Divita, students assembled all the ingredients of the DNA origami structure and followed a procedure to anneal the DNA origami structure. Visualization of our DNA origami structure by Atomic Force Microscopy (AFM) was conducted by Divita. Students observed and interacted with Divita during the imaging process. Earlier in the month we visited with the developers at Novascan who made our instrument where we learned new AFM techniques which we have already put into use.
These visits provided us with hands-on experience in practical techniques for constructing and visualizing DNA origami structures. All of our six students were given the opportunity to implement the best known practices in DNA origami research. We have refined our techniques for DNA origami construction and atomic force microscope visualization and will apply these techniques to advance our research capabilities at Simpson College over the following months. We are planning to present the results from this project next fall at the DNA 19 conference in Arizona.